1 Modeling¶
1.1 Import Requisite Libraries¶
In [1]:
######################## Standard Library Imports ##############################
import pandas as pd
import numpy as np
import os
import sys
from eda_toolkit import ensure_directory, generate_table1
######################## Modeling Library Imports ##############################
import shap
from model_tuner.pickleObjects import loadObjects
import model_tuner
import eda_toolkit
from sklearn.decomposition import PCA
from sklearn.preprocessing import LabelEncoder
import matplotlib.pyplot as plt
# Add the parent directory to sys.path to access 'functions.py'
sys.path.append(os.path.join(os.pardir))
print(
f"This project uses: \n \n Python {sys.version.split()[0]} \n model_tuner "
f"{model_tuner.__version__} \n eda_toolkit {eda_toolkit.__version__}"
)
1.2 Set Paths & Read in the Data¶
In [2]:
# Define your base paths
# `base_path`` represents the parent directory of your current working directory
base_path = os.path.join(os.pardir)
# Go up one level from 'notebooks' to the parent directory, then into the 'data' folder
data_path = os.path.join(os.pardir, "data")
image_path_png = os.path.join(base_path, "images", "png_images", "modeling")
image_path_svg = os.path.join(base_path, "images", "svg_images", "modeling")
# Use the function to ensure the 'data' directory exists
ensure_directory(data_path)
ensure_directory(image_path_png)
ensure_directory(image_path_svg)
In [3]:
data_path = "../data/processed/"
model_path = "../mlruns/models/"
In [4]:
df = pd.read_parquet(os.path.join(data_path, "X.parquet"))
In [5]:
df.columns.to_list()
Out[5]:
In [6]:
X = pd.read_parquet(os.path.join(data_path, "X.parquet"))
y = pd.read_parquet(os.path.join(data_path, "y_Bleeding_Edema_Outcome.parquet"))
df = df.join(y, how="inner", on="patient_id")
1.2.1 Load Models¶
In [7]:
# lr_smote_training
model_lr = loadObjects(
os.path.join(
model_path,
"./452642104975561062/8eab72fdaa134c209521879f18f19d06/artifacts/lr_Bleeding_Edema_Outcome/model.pkl",
)
)
# rf_over_training
model_rf = loadObjects(
os.path.join(
model_path,
"./452642104975561062/d18ee7233d0f40ae968e57b596b75ac7/artifacts/rf_Bleeding_Edema_Outcome/model.pkl",
)
)
# svm_orig_training
model_svm = loadObjects(
os.path.join(
model_path,
"./452642104975561062/18dc58511b9e45ebaf55308026701c18/artifacts/svm_Bleeding_Edema_Outcome/model.pkl",
)
)
1.2.2 Set-up Pipelines, Model Titles, and Thresholds¶
In [8]:
pipelines_or_models = [model_lr, model_rf, model_svm]
# Model titles
model_titles = [
"Logistic Regression",
"Random Forest Classifier",
"Support Vector Machines",
]
thresholds = {
"Logistic Regression": next(iter(model_lr.threshold.values())),
"Random Forest Classifier": next(iter(model_rf.threshold.values())),
"Support Vector Machines": next(iter(model_svm.threshold.values())),
}
In [9]:
for col in X.columns:
if col.startswith("BMI_"):
print(f"Value Counts for column {col}:\n")
print(X[col].value_counts())
print("\n")
1.3 Summarize Model Performance¶
In [10]:
from model_metrics import summarize_model_performance
table3 = summarize_model_performance(
model=pipelines_or_models,
X=X,
y=y,
model_title=model_titles,
model_threshold=thresholds,
return_df=True,
)
In [11]:
table3
Out[11]:
1.4 SHAP Summary Plot¶
1.4.1 SHAP (SHapley Additive exPlanations) Set-up¶
In [12]:
# Step 1: Get transformed features using model's preprocessing pipeline
X_transformed = model_svm.get_preprocessing_and_feature_selection_pipeline().transform(
X
)
# Optional: Sampling for speed (or just use X_transformed if it's small)
sample_size = 100
X_sample = shap.utils.sample(X_transformed, sample_size, random_state=42)
# Step 2: Get final fitted model (SVC in your pipeline)
final_model = model_svm.estimator.named_steps[model_svm.estimator_name]
# Step 3: Define a pred. function that returns only the probability for class 1
def model_predict(X):
return final_model.predict_proba(X)[:, 1]
# Step 4: Create SHAP explainer
explainer = shap.KernelExplainer(
model_predict, X_sample, feature_names=model_svm.get_feature_names()
)
# Step 5: Compute SHAP values for the full dataset or sample
shap_values = explainer.shap_values(X_sample) # can use X_transformed instead
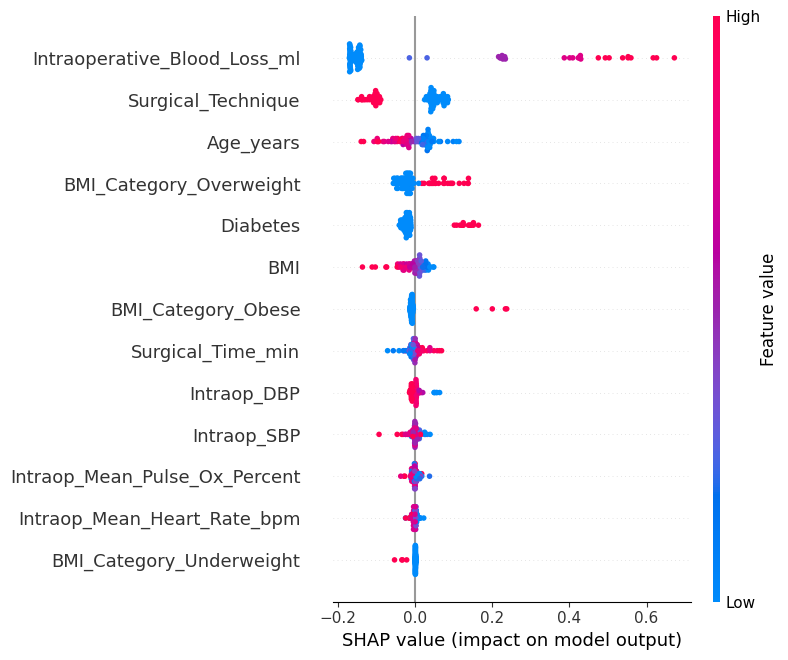
1.4.2 SHAP Beeswarm Plot¶
In [13]:
# Step 6a: SHAP beeswarm plot (default)
shap.summary_plot(
shap_values,
X_sample,
feature_names=model_svm.get_feature_names(),
show=False,
)
plt.savefig(os.path.join(image_path_png, "shap_summary_beeswarm.png"), dpi=600)
plt.savefig(os.path.join(image_path_svg, "shap_summary_beeswarm.png"), dpi=600)
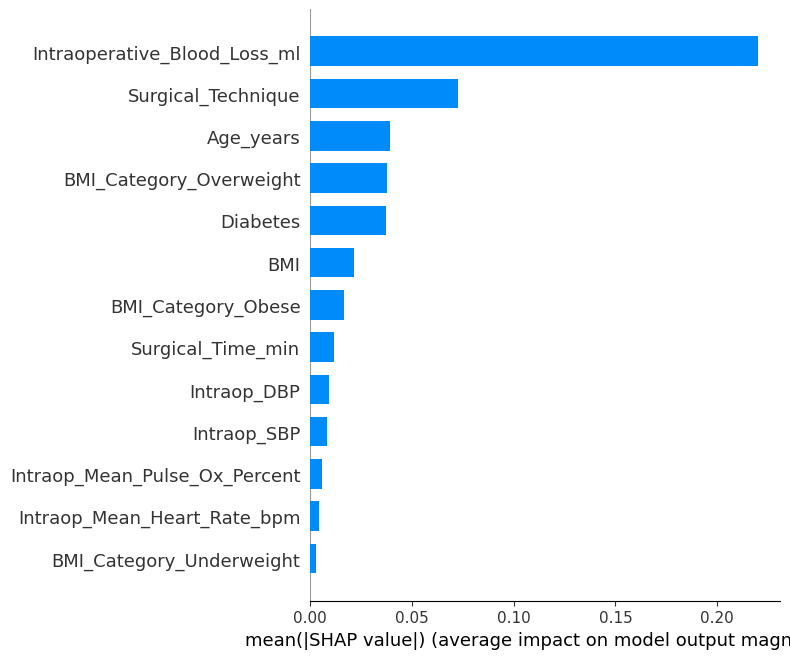
1.4.3 SHAP Bar Plot¶
In [14]:
# Step 6b: SHAP bar plot (mean |SHAP value| for each feature)
shap.summary_plot(
shap_values,
X_sample,
feature_names=model_svm.get_feature_names(),
plot_type="bar",
show=False,
)
plt.savefig(os.path.join(image_path_png, "shap_summary_bar.png"), dpi=600)
plt.savefig(os.path.join(image_path_svg, "shap_summary_bar.png"), dpi=600)
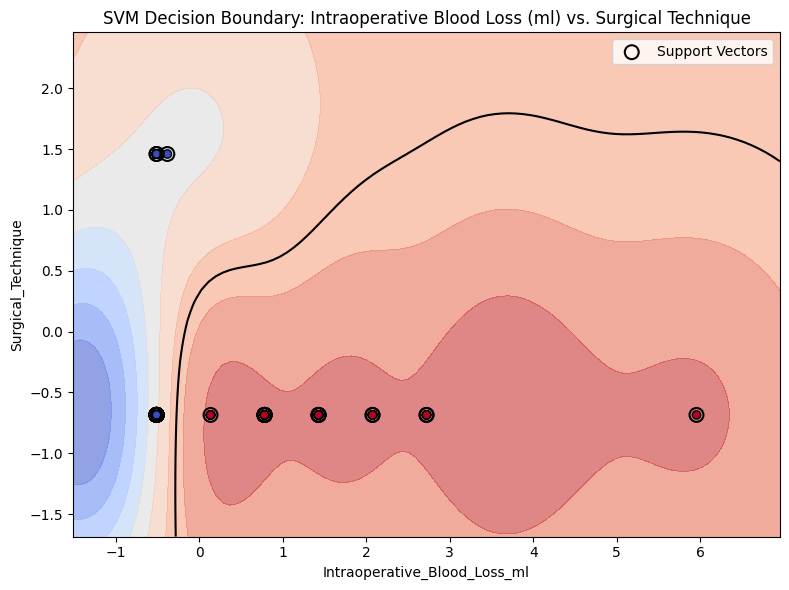
1.5 Plot SVM Decision Boundary¶
In [15]:
from project_functions import plot_svm_decision_boundary_2d
plot_svm_decision_boundary_2d(
# model=model_svm,
X=X,
y=y,
feature_pair=("Intraoperative_Blood_Loss_ml", "Surgical_Technique"),
title="SVM Decision Boundary: Intraoperative Blood Loss (ml) vs. Surgical Technique",
image_path_svg=os.path.join(image_path_svg, "svm_decision_surface_2d.svg"),
)
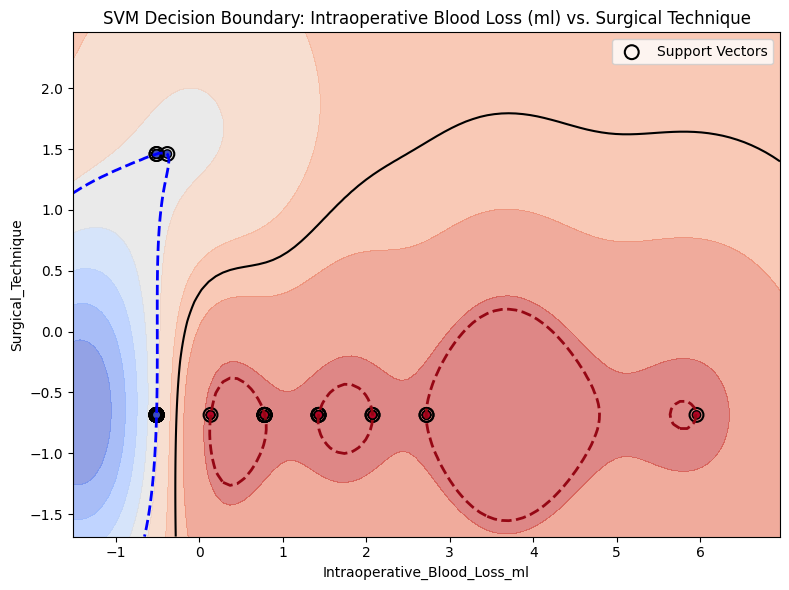
In [16]:
from project_functions import plot_svm_decision_boundary_2d
plot_svm_decision_boundary_2d(
# model=model_svm,
X=X,
y=y,
feature_pair=("Intraoperative_Blood_Loss_ml", "Surgical_Technique"),
title="SVM Decision Boundary: Intraoperative Blood Loss (ml) vs. Surgical Technique",
margin=True,
image_path_svg=os.path.join(image_path_svg, "svm_decision_surface_2d_margin.svg"),
)
In [17]:
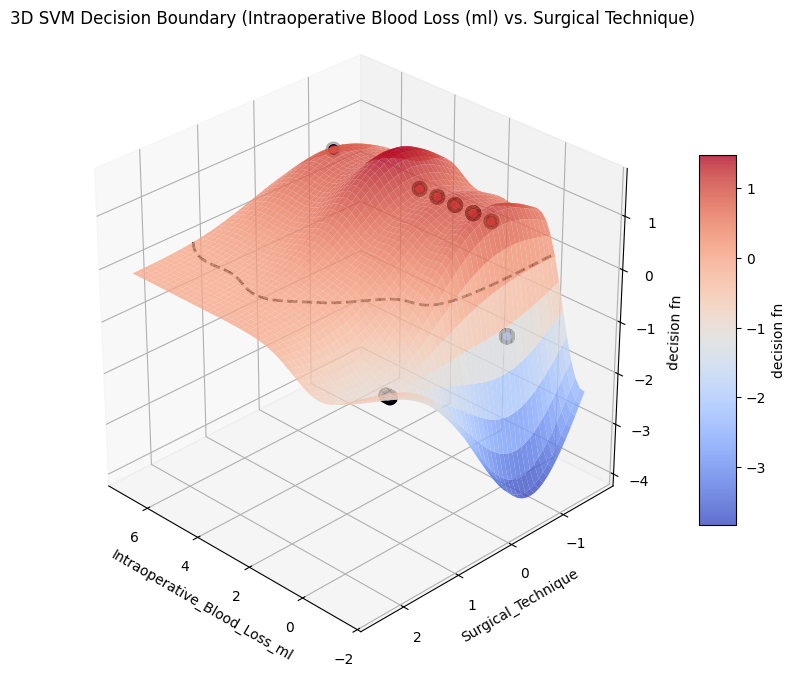
from project_functions import plot_svm_decision_surface_3d
plot_svm_decision_surface_3d(
X=X,
y=y,
# figsize=(6, 10),
feature_pair=("Intraoperative_Blood_Loss_ml", "Surgical_Technique"),
title="3D SVM Decision Boundary (Intraoperative Blood Loss (ml) vs. Surgical Technique)",
image_path_png=os.path.join(image_path_png, "svm_decision_surface_3d.png"),
image_path_svg=os.path.join(image_path_svg, "svm_decision_surface_3d.svg"),
)
In [18]:
from project_functions import plot_svm_decision_surface_3d_plotly
# Plotly 3D SVM Decision Surface
plot_svm_decision_surface_3d_plotly(
X=df,
y=df["Bleeding_Edema_Outcome"],
feature_pair=("Intraoperative_Blood_Loss_ml", "Surgical_Technique"),
title="Interactive 3D SVM Decision Boundary: <br> Intraoperative Blood Loss (ml) vs. Surgical Technique",
html_path=os.path.join(image_path_svg, "svm_decision_surface_3d_plotly.html"),
)
1.6 Calibration¶
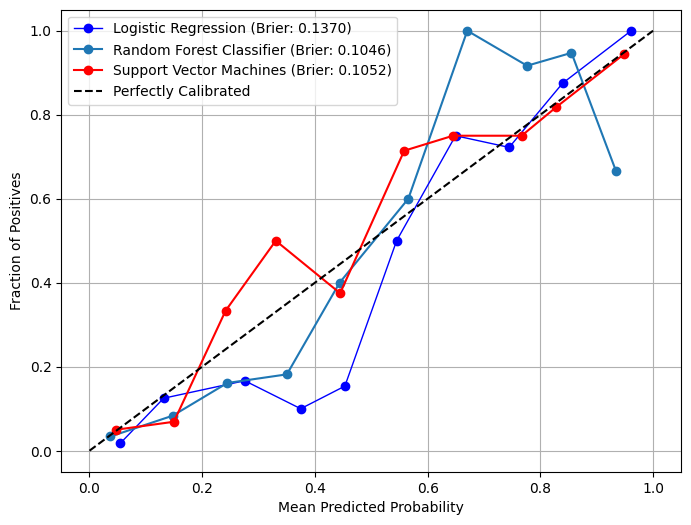
In [19]:
# Plot calibration curves in overlay mode
from model_metrics import show_calibration_curve
show_calibration_curve(
model=pipelines_or_models,
X=X,
y=y,
model_title=model_titles,
overlay=True,
title="",
save_plot=True,
image_path_png=image_path_png,
image_path_svg=image_path_svg,
text_wrap=40,
curve_kwgs={
"Logistic Regression": {"color": "blue", "linewidth": 1},
"Support Vector Machines": {
"color": "red",
# "linestyle": "--",
"linewidth": 1.5,
},
"Decision Tree": {
"color": "lightblue",
"linestyle": "--",
"linewidth": 1.5,
},
},
figsize=(8, 6),
label_fontsize=10,
tick_fontsize=10,
bins=10,
show_brier_score=True,
grid=False,
# gridlines=False,
linestyle_kwgs={"color": "black"},
dpi=500,
)
1.7 Confusion Matrices¶
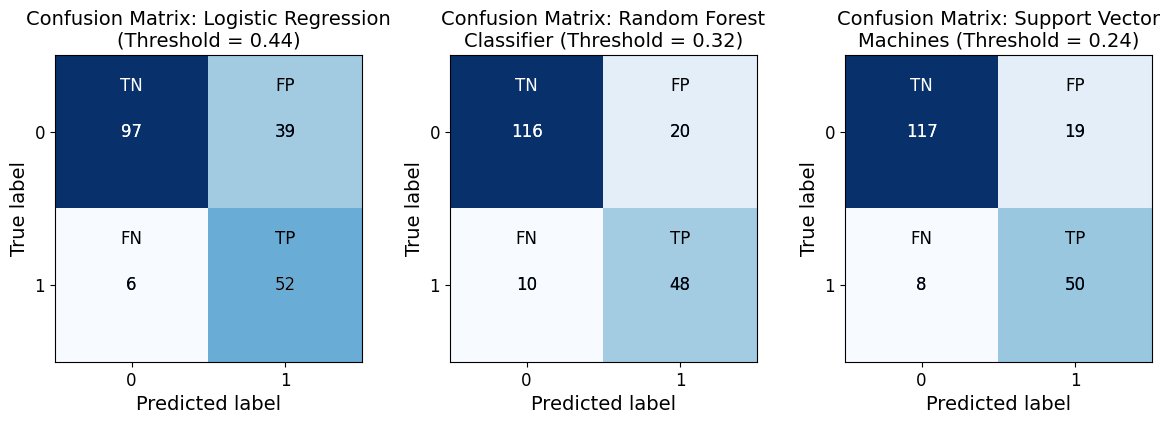
In [20]:
from model_metrics import show_confusion_matrix
show_confusion_matrix(
model=pipelines_or_models,
X=X,
y=y,
model_title=model_titles,
model_threshold=[thresholds],
# class_labels=["No Pain", "Class 1"],
cmap="Blues",
text_wrap=40,
save_plot=True,
image_path_png=image_path_png,
image_path_svg=image_path_svg,
grid=True,
n_cols=3,
n_rows=1,
figsize=(4, 4),
show_colorbar=False,
label_fontsize=14,
tick_fontsize=12,
inner_fontsize=12,
class_report=True,
# thresholds=thresholds,
# custom_threshold=0.5,
# labels=False,
)
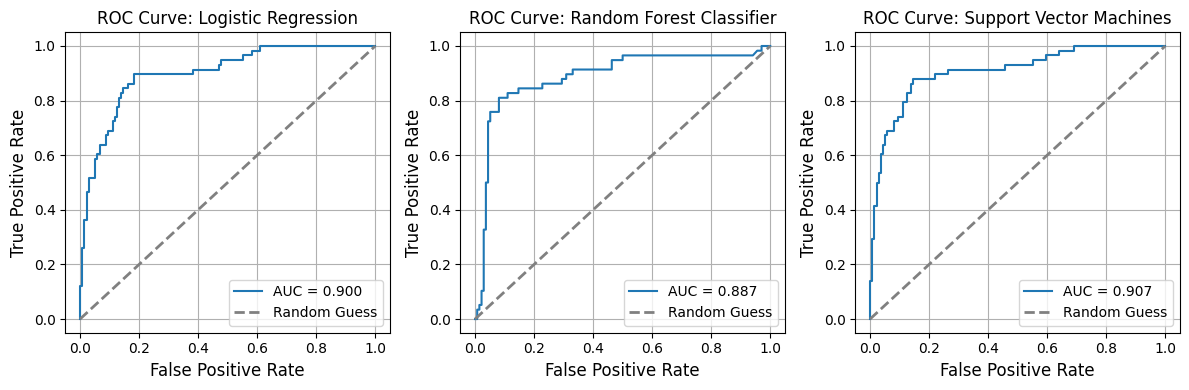
1.8 ROC AUC Curves¶
In [21]:
from model_metrics import show_roc_curve
# Plot ROC curves
show_roc_curve(
model=pipelines_or_models,
X=X,
y=y,
overlay=False,
model_title=model_titles,
decimal_places=3,
# n_cols=3,
# n_rows=1,
# curve_kwgs={
# "Logistic Regression": {"color": "blue", "linewidth": 2},
# "SVM": {"color": "red", "linestyle": "--", "linewidth": 1.5},
# },
# linestyle_kwgs={"color": "grey", "linestyle": "--"},
save_plot=True,
grid=True,
n_cols=3,
figsize=(12, 4),
# label_fontsize=16,
# tick_fontsize=16,
image_path_png=image_path_png,
image_path_svg=image_path_svg,
# gridlines=False,
)
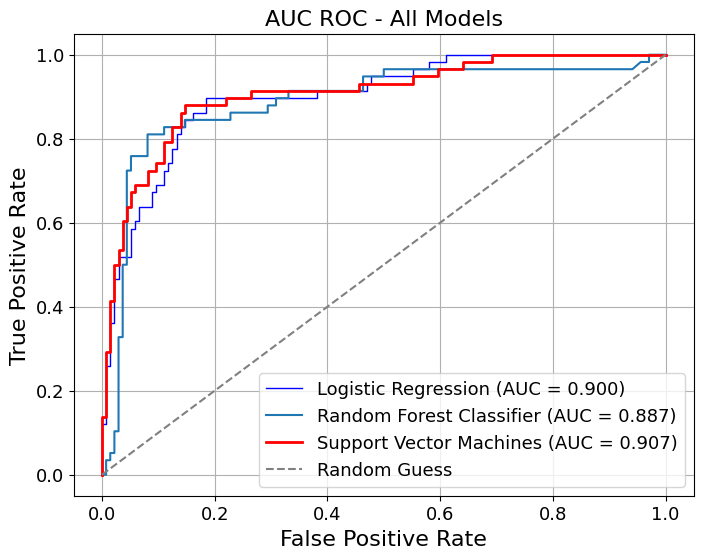
In [22]:
show_roc_curve(
model=pipelines_or_models,
X=X,
y=y,
overlay=True,
model_title=model_titles,
title="AUC ROC - All Models",
curve_kwgs={
"Logistic Regression": {"color": "blue", "linewidth": 1},
"Random Forest": {"color": "lightblue", "linewidth": 1},
"Support Vector Machines": {
"color": "red",
"linestyle": "-",
"linewidth": 2,
},
},
linestyle_kwgs={"color": "grey", "linestyle": "--"},
save_plot=True,
grid=False,
decimal_places=3,
figsize=(8, 6),
# gridlines=False,
label_fontsize=16,
tick_fontsize=13,
image_path_png=image_path_png,
image_path_svg=image_path_svg,
dpi=500,
)
1.9 Precision-Recall Curves¶
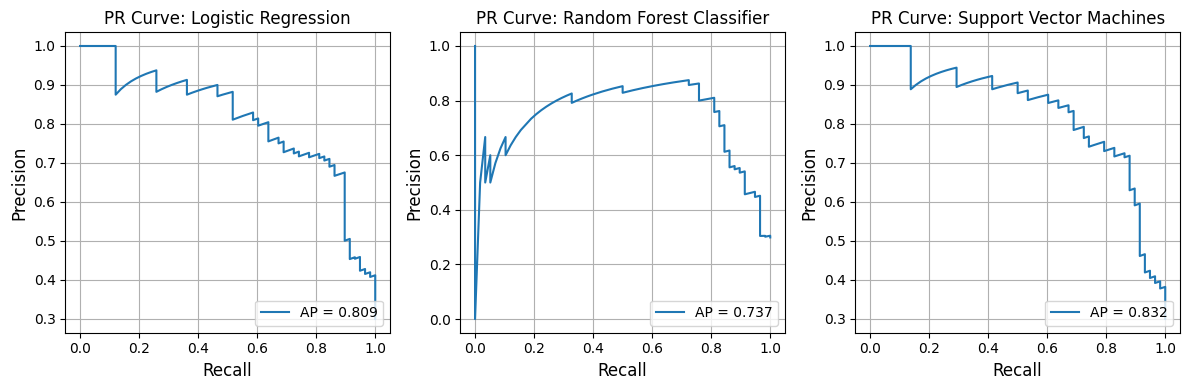
In [23]:
from model_metrics import show_pr_curve
# Plot PR curves
show_pr_curve(
model=pipelines_or_models,
X=X,
y=y,
# x_label="Hello",
model_title=model_titles,
decimal_places=3,
overlay=False,
grid=True,
save_plot=True,
image_path_png=image_path_png,
image_path_svg=image_path_svg,
figsize=(12, 4),
n_cols=3,
# tick_fontsize=16,
# label_fontsize=16,
# grid=True,
# gridlines=False,
)
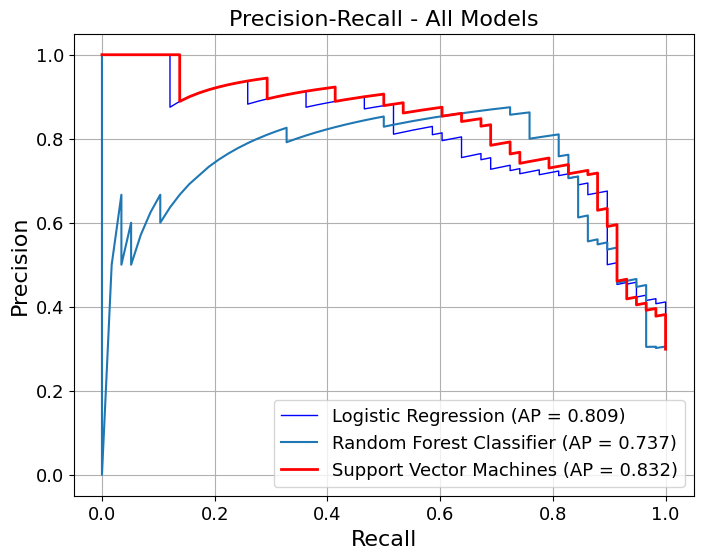
In [24]:
show_pr_curve(
model=pipelines_or_models,
X=X,
y=y,
overlay=True,
model_title=model_titles,
title="Precision-Recall - All Models",
curve_kwgs={
"Logistic Regression": {"color": "blue", "linewidth": 1},
"Random Forest": {"color": "lightblue", "linewidth": 1},
"Support Vector Machines": {
"color": "red",
"linestyle": "-",
"linewidth": 2,
},
},
save_plot=True,
grid=False,
decimal_places=3,
figsize=(8, 6),
# gridlines=False,
label_fontsize=16,
tick_fontsize=13,
image_path_png=image_path_png,
image_path_svg=image_path_svg,
dpi=500,
)